Data Science and Machine Learning Internship ...
- 22k Enrolled Learners
- Weekend/Weekday
- Live Class
R for Data Science is a must learn for Data Analysis & Data Science professionals. With its growth in the IT industry, there is a booming demand for skilled Data Scientists who have an understanding of the major concepts in R. One such concept, is the Decision Tree.
In this blog we will discuss :
1. How to create a decision tree for the admission data.
2. Use rattle to plot the tree.
3. Validation of decision tree using the ‘Complexity Parameter’ and cross validated error.
4. Prune the tree on the basis of these parameters to create an optimal decision tree.
To understand what are decision trees and what is the statistical mechanism behind them, you can read this post : How To Create A Perfect Decision Tree
To create a decision tree in R, we need to make use of the functions rpart(), or tree(), party(), etc.
rpart() package is used to create the tree. It allows us to grow the whole tree using all the attributes present in the data.
> library("rpart")
> setwd("D://Data")
> data <- read.csv("Gre_Coll_Adm.csv")
> str(data)
'data.frame': 400 obs. of 5 variables:
$ X : int 1 2 3 4 5 6 7 8 9 10 ...
$ Admission_YN : int 0 1 1 1 0 1 1 0 1 0 ...
$ Grad_Rec_Exam: int 380 660 800 640 520 760 560 400 540 700 ...
$ Grad_Per : num 3.61 3.67 4 3.19 2.93 3 2.98 3.08 3.39 3.92 ...
$ Rank_of_col : int 3 3 1 4 4 2 1 2 3 2 ...
> View(data)> adm_data<-as.data.frame(data) > tree <- rpart(Admission_YN ~ adm_data$Grad_Rec_Exam + adm_data$Grad_Per+ adm_data$Rank_of_col, + data=adm_data, + method="class")
rpart syntax takes ‘dependent attribute’ and the rest of the attributes are independent in the analysis.
Admission_YN : Dependent Attribute. As admission depends on the factors score, rank of college, etc.
Grad_Rec_Exam, Grad_Per, and Rank_of_col : Independent Attributes
rpart() returns a Decison tree created for the data.
If you plot this tree, you can see that it is not visible, due to the limitations of the plot window in the R console.
> plot(tree) > text(tree, pretty=0)
To enhance it, let us take some help from rattle :
> library(rattle) > rattle()
Rattle() is one unique feature of R which is specifically built for data mining in R. It provides its own GUI apart from the R Console which makes it easier to analyze data. It has built-in graphics, which provides us better visualizations as well. Here we will use just the plotting capabilities of Rattle to achieve a decent decision tree plot.
> library(rpart.plot) > library(RColorBrewer)
rpart.plot() and RcolorBrewer() functions help us to create a beautiful plot. ‘rpart.plot()’ plots rpart models. It extends plot.rpart and text.rpart in the rpart package. RcolorBrewer() provides us with beautiful color palettes and graphics for the plots.
> fancyRpartPlot(tree)
This was a simple and efficient way to create a Decision Tree in R. But are you sure that this is the optimal ‘Decision Tree’ for this data? If not, the following validation checks will help you.
Meanwhile, if you wish to learn R programming, check out our specially curated course by clicking on the below button.
To validate the model we use the printcp and plotcp functions. ‘CP’ stands for Complexity Parameter of the tree.
Syntax : printcp ( x ) where x is the rpart object.
This function provides the optimal prunings based on the cp value.
We prune the tree to avoid any overfitting of the data. The convention is to have a small tree and the one with least cross validated error given by printcp() function i.e. ‘xerror’.
Cross Validated Error :
To find out how the tree performs, is calculated by the printcp() function, based on which we can go ahead and prune the tree.
> printcp(tree) Classification tree: rpart(formula = Admission_YN ~ adm_data$Grad_Rec_Exam + adm_data$Grad_Per + adm_data$Rank_of_col, data = adm_data, method = "class") Variables actually used in tree construction: [1] adm_data$Grad_Per adm_data$Grad_Rec_Exam adm_data$Rank_of_col Root node error: 127/400 = 0.3175 n= 400 CP nsplit rel error xerror xstd 1 0.062992 0 1.00000 1.00000 0.073308 2 0.023622 2 0.87402 0.92913 0.071818 3 0.015748 4 0.82677 0.99213 0.073152 4 0.010000 8 0.76378 1.02362 0.073760
From the above mentioned list of cp values, we can select the one having the least cross-validated error and use it to prune the tree.
The value of cp should be least, so that the cross-validated error rate is minimum.
To select this, you can make use of this :
fit$cptable[which.min(fit$cptable[,”xerror”]),”CP”]
This function returns the optimal cp value associated with the minimum error.
Let us see what plotcp() function fetches.
> plotcp(tree)
Plotcp() provides a graphical representation to the cross validated error summary. The cp values are plotted against the geometric mean to depict the deviation until the minimum value is reached.
> ptree<- prune(tree, + cp= tree$cptable[which.min(tree$cptable[,"xerror"]),"CP"]) > fancyRpartPlot(ptree, uniform=TRUE, + main="Pruned Classification Tree")
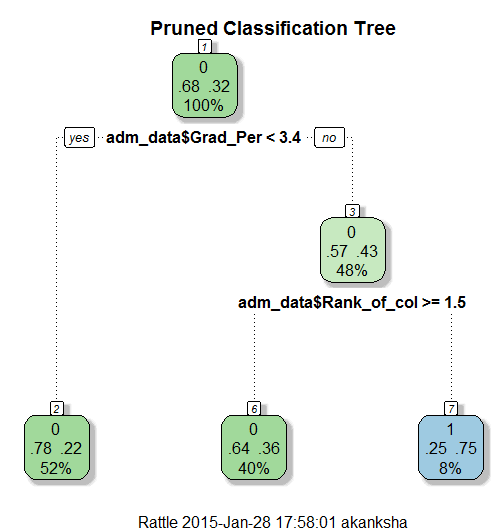
Thus we create a pruned decision tree.
If you wish to get a head-start on R programming, check out the Data Analytics with R course from Edureka.
Got a question for us? Please mention them in the comments section and we will get back to you.
Related Posts:
 Thank you for registering Join Edureka Meetup community for 100+ Free Webinars each month JOIN MEETUP GROUP
Thank you for registering Join Edureka Meetup community for 100+ Free Webinars each month JOIN MEETUP GROUPedureka.co
Hi Team,
Nice explanation. Can you please share the dataset used here to my mail id:achyut.k@gmail.com
hey! Can you send the data set to me please :) mikeyling18@gmail.com
Hi. Can you share the dataset? My email is patneshubham123@gmail.com. Thanks!
Can you send the data set with me at parmarpraveen18@gmail.com
HI dear,
Thanks for this article. Please, can you share your data set with me (nadege.agandan@gmail.com). Thanks a bunch
Hey Agandan, we have shared the files with you. Do let us know if you need anything else. We hoped you liked our content! :)
Hi can you shared the the files with me my mail es carlosalbuquerquec@gmail.com
Hi. Can you share the dataset? My email is erfavaro@gmail.com. Thanks!
Hey Edio, we have shared the files with you. Do let us know if you need anything else. We hoped you liked our content! :)